Push and pull: how to measure the forces that sculpt embryos

A steadily growing toolbox is giving researchers the ability to monitor and measure the physical forces that shape embryonic development
Embryonic development is like an elaborate stage production, in which meticulously choreographed gene-expression programmes organize much of the action. But changes in physical forces such as tension and mechanical properties like elasticity also play a part, coordinating the process’s spatial and temporal footwork.
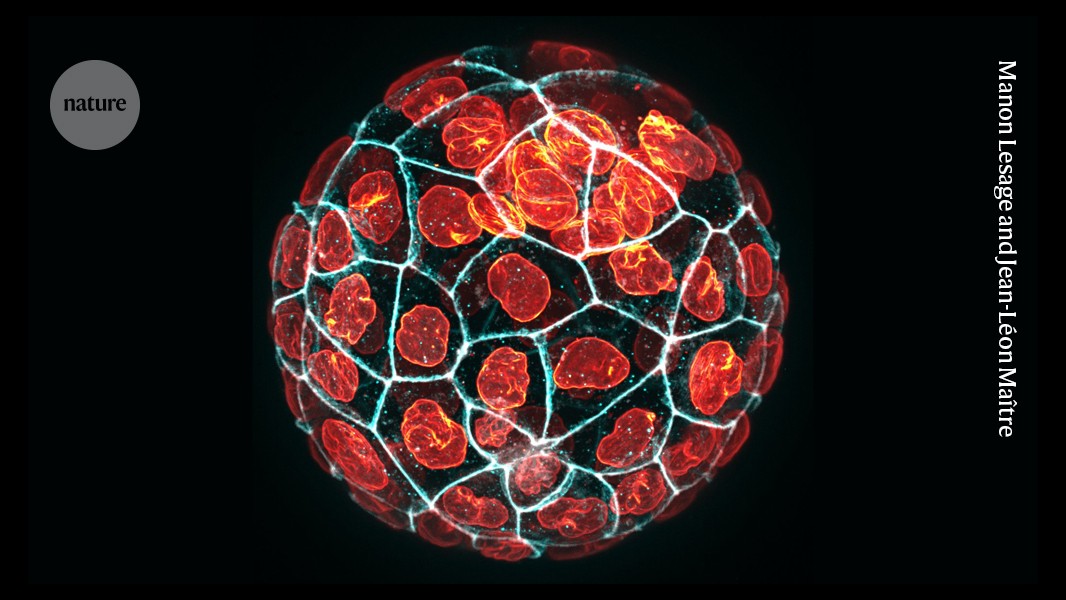
“Mechanical properties actually become relevant to formation of the first cell lineages before gene expression,” says Jean-Léon Maître, a mechanobiologist at the Curie Institute in Paris. After fertilization, subsequent rounds of cell division produce extensive rearrangements and deformations. The resulting squeezing, stretching, pushing and pulling influences which cells form different embryonic features, and ultimately guides the patterning and development of every tissue in the body. “You first have this difference in mechanics, which then translates to a difference in position, which then translates into fate,” says Maître.
Measuring these properties and how they change over time is therefore essential to understanding embryonic development. But making such observations isn’t easy. Many biophysical and mechanobiology techniques are best suited to artificial systems with cultured cells. Promising techniques are emerging for in vivo studies, but these require further testing to prove their accuracy and quantitative mettle. “All of it is still relatively hard,” says Amy Shyer, a mechanobiologist who co-directs a laboratory at Rockefeller University in New York City with developmental biologist Alan Rodrigues. “There isn’t an off-the-shelf toolkit.” The challenges only mount with live embryos, in which the goal is to measure physical and mechanical phenomena without disrupting development.
But a steady rise in interest in mechanobiology is leading to technologies that offer exciting opportunities to establish a more holistic view of developmental biology. “We’re just at that moment where the field can take off,” says Otger Campàs, a biological physicist at the Technical University Dresden in Germany.
Cells in living tissues have multiple levers that they can pull to reorganize themselves and physically influence their neighbours. The cytoskeleton is a network of proteins that helps to define the shape and organization of cells. By rearranging that internal structure, cells can push and pull and even travel over or between one another, and alter their own mechanical properties, transitioning between being squishy and fluid or firm and viscous.
Under pressure
Furthermore, cell division can contribute to crowding and create changes in embryonic surface tension that are especially important early in development. “To position these cells in a certain place, you need to actually control the way they divide,” says Hervé Turlier, a biophysicist at the College of France in Paris. The early embryo, he notes, starts as a seemingly homogeneous ball of cells; the directionality of cell division helps to disrupt this symmetry and establish the axes that determine an organism’s front, back, top and bottom. One 2016 study, for instance, showed that inward-directed forces associated with certain early cell-division events help to separate cells that form the mouse embryo from those that give rise to extra-embryonic tissues such as the yolk sac1.
Which method is chosen to investigate these processes depends, in part, on the experimental model being studied. Mammalian embryos, such as mice, can be challenging because they generally cannot be maintained outside the mother’s body beyond the stage at which uterine implantation occurs, roughly four days post-fertilization. Moreover, each cell-division event takes place over many hours, requiring long-term experiments. Model organisms such as the roundworm Caenorhabditis elegans offer a simpler alternative. But with their ultrafast development and cycles of cell division that are separated by minutes rather than hours, some methods have trouble keeping up. This frenetic pace also introduces frictional forces that are absent in more slowly dividing embryos, Maître says, making it challenging to model the organism’s mechanical properties.
The good news, says developmental biologist Carl-Philipp Heisenberg at the Institute of Science and Technology Austria in Klosterneuburg, is that many fundamental mechanisms are conserved across species, at least in early development. “The general principles you’re looking at — how cells are interacting, how mechanical forces are being generated, transmitted and sensed — they’re being used again and again in different organisms,” he says.
Full contact
Certain mechanobiology tools are also being used again and again, and two in particular have been popular for decades.
Atomic force microscopy (AFM) was developed in the 1980s as a way to probe the surface texture and stiffness of materials with atomic-scale precision. AFM uses a tiny, cantilevered probe, like a diving board, that is dragged across a sample surface and illuminated with a laser. Changes in the reflected light can be used to calculate tissue stiffness and elasticity, as well as the force with which two cells are attached.
The other approach, micropipette aspiration, dates to the 1950s, when it was used to study sea urchin embryos. Researchers place the tip of an extremely fine glass pipette against a cell surface, then use a pump to draw the membrane part way into the pipette to test membrane surface tension and cell viscosity, or to push air against it to apply a controlled force. With multiple pipettes, researchers can play ‘tug of war’ with adjoining cells to determine how tightly they are coupled. The result is an absolute, rather than relative, measure of force, says Chii Jou Chan, a mechanobiologist at the National University of Singapore.
Jean-Léon Maître, who uses micropipette aspiration to map surface tension of embryos.Credit: Alexandre Darmon/Art in Research for the Bettencourt Schueller Foundation
Maître’s lab has used micropipette aspiration to extensively map surface tension in the earliest stages of mouse development, when embryos are still relatively simple balls of cells. In a study published in May, he and his colleagues applied the technique to study compaction — the process by which early embryos establish tighter couplings between cells — in human embryos2. The researchers identified mechanical properties that could inform the selection of viable embryos for reproductive technologies, such as in vitro fertilization.
AFM, in contrast, has limited utility in live embryos. “You need something flat,” explains Shyer. But the technique can deliver important insights when applied to tissues that have been removed from developing embryos and studied in culture. In 2023 study, Shyer and Rodrigues used techniques including AFM and micropipette aspiration to measure how different morphogens — molecules that act as developmental signals —affect the mechanical properties of developing skin and follicles in chicken embryos3. They found that the morphogen-induced biophysical changes that precede follicle formation occur not at the single-cell level as was previously thought, but on the scale of large cell groups. “It was sort of the first time measuring emergent physical properties at this collective cell scale,” says Shyer, noting that most biomechanical studies focus on individual cells.
That said, many aspects of development can be explored only with whole embryos. “If we care about the physics of how embryonic structures are formed, then we need to measure physical fields inside these structures as they’re being formed,” says Campàs.
Inside perspective
The toolbox for such work has been limited, however. Alongside micropipette aspiration, for instance, many researchers evaluated surface tension by using ultraviolet lasers to introduce targeted cuts on the embryo surface. “You observe sort of a recoil of the tissue which you have cut, and this recoil velocity is proportional to the tension in this tissue,” explains Heisenberg. But the embryo is damaged in the process, and the results are difficult to quantify because the strength of recoil is inherently dependent on the — potentially unknown — mechanical properties of the affected cells.
Now, other promising methods are emerging. The Campàs lab has developed oil-encapsulated droplet sensors that can be injected into live embryos and then imaged. In the original iteration of these sensors, the droplets are filled with fluorescent dye, and the stresses they experience at a given injection site are calculated on the basis of how much they deform. This approach has proven especially valuable in fast-developing models such as zebrafish. “You can image an entire organ forming in just a few hours,” says Campàs. But it is also suitable for mammalian development. In 2022, for instance, Campàs and his colleagues used this approach to document how the gradual build-up of internal physical stress governs the initiation and extent of toe growth in embryonic mice4. Mutant mice that lack a morphogen that contributes to this stress build-up, developed only short nubs rather than functional digits.

A droplet (pink and cyan) in between cells of a living zebrafish embryo allows scientists to measure the osmotic pressure in the tissue.Credit: A. Vian et al./Nature Commun. (CC BY 4.0)
Campàs’s team has also developed more sophisticated microdroplet sensors. One version incorporates an iron-based ‘ferrofluid’ that can be manipulated with a magnetic field5. This enables researchers to apply controlled forces at the injection site for extended periods while also measuring the material properties of the surrounding tissue. Another iteration features a water droplet encased in an oil droplet6. When this droplet in a droplet is subject to increased osmotic pressure — for example, if a cell or tissue becomes saturated — it takes up more fluid, producing a measurable change in volume. Campàs says that these sensors could be useful for understanding how channels form in hollow organ structures, such as the airways of the lung or in pancreatic ducts.
Optical tweezers, which use focused lasers to physically ‘trap’ and manipulate nanoscale objects, can also be used to push and pull cellular membranes and organelles in vivo. Pierre-François Lenne at Aix Marseille University in France has used this technology to test the strength of cell–cell junctions in live fruit fly embryos. “He traps the junction, and then he would basically just play the guitar with it,” says Maître. Such experiments have historically required complex, home-built apparatus, but commercial platforms are now available, and Maître is using the method in his own lab to test how the material properties of cell and nuclear membranes change as embryos grow. “It’s a very promising tool,” he says.
A hands-off approach
That said, the same sensing mechanisms that allow cells to follow force-induced prompts can also react to being poked with a pipette tip, Heisenberg cautions, and even relatively gentle interventions such as the injection of microdroplets could elicit a response. “You’re putting it into a tissue, and the tissue knows that there is something [there] which doesn’t belong,” says Heisenberg.
Researchers are, therefore, keen to develop contact-free alternatives. One possibility is to computationally deduce the forces that individual cells experience on the basis of how much they deform at junctions with other cells. “The surface is what mostly controls the shape of cells,” explains Turlier. “It’s not things pushing from inside, it’s really the surface that deforms itself.” Given sufficient starting information, researchers could, in principle, map the force landscape of an embryo from image analysis alone.

Foambryo software models embryos as foams of bubble-like cells.Credit: Collège de France
Many tools for force inference are best suited to flat sheets of epithelial cells, but Turlier’s group has been making headway in 3D force inference using an algorithm called ‘foambryo’7. Published in 2023, foambryo models embryos as foams of bubble-like cells, an assumption that seems to be broadly applicable across embryonic systems in terms of the general shape and arrangement of cells. Foambryo has important limitations, Turlier notes — for example, the method provides relative rather than absolute force measurements, and accounts for only a subset of the tensions that embryonic cells are subject to. Still, he says, “I think what we’ve done here is important, because even if it’s not perfect, it will give us a good first guess.”
In another emerging non-invasive method, Brillouin microscopy, samples are scanned with a laser beam in a modified fluorescence microscope. The movement of biomolecules in the scanned tissue influences the extent to which that light is scattered, and measurements of this optical scatter can then be used to calculate mechanical properties associated with cellular stiffness and elasticity.
Jitao Zhang, a biomedical engineer at Wayne State University in Detroit, Michigan, is among the method’s champions and has been using Brillouin microscopy to study essential processes in development that can contribute to life-threatening congenital conditions if derailed, such as spina bifida. “We’ve applied this optical method to chick embryos to monitor how the stiffness changes when the neural tube is closing,” says Zhang. Brillouin allowed his team to document this process over more than 21 hours, measuring a steady increase in tissue stiffness as the neural-tube tissue thickens, bends and closes8. Chan has also found Brillouin microscopy to be a valuable tool. “I think it’s a perfect system to study intrafollicular mechanical stiffness,” he says.
Nevertheless, there are open questions about the utility of Brillouin microscopy. The mechanical measurements it enables are influenced by the refractive index and density of the tissue being imaged, and these parameters might not be well defined. “It’s based on assumptions which are sometimes experimentally very difficult to prove,” says Heisenberg. Methods are emerging that make it possible to measure the optical properties of complex biological samples, Chan notes. And although AFM and Brillouin measure different mechanical properties, the two seem to correlate in many biological systems. Cross-validation is therefore important for interpreting Brillouin data, and Zhang says that his group is careful to fact-check its Brillouin data using AFM. “You have to do sample-dependent calibration — that’s the painful part,” he says. His team is setting up a dedicated facility in which both methods can be carried out on the same sample in parallel.
Filling in the gaps
Indeed, when it comes to mechanobiology, even the most tried-and-tested methods present uncertainties. “You’re dealing with a living material,” says Rodrigues. “There’s a little bit of trickiness there because you have to try to build tools about a substance that you don’t fully even understand.”
That means any study will require assumptions and models that are based on incomplete information about parameters that might vary considerably between different embryos and over time. For example, Turlier notes that incorrect assumptions about embryonic cell geometry have led to widely divergent results using AFM. It’s not unheard of, he says, for repeated measurements on the same cell to differ by as much as 700%. More in vivo data should prove valuable on this front, he adds, generating values that can be used to build and calibrate more accurate models.
And then there’s the challenge of tying these mechanical phenomena back to the underlying genetic and biomolecular processes that elicit them. Studies like those of Shyer and Rodrigues in avian skin demonstrate the feasibility of drawing explicit connections between specific morphogens or other signalling prompts and large-scale changes in mechanical properties and force maps. Rodrigues is enthusiastic about the opportunity to close the circle here, and perhaps derive more comprehensive explanations for the root causes of poorly understood developmental disorders. “Understanding biophysics at this level”, he says, “could allow us to actually make sense of a lot of genetic information that we haven’t been able to organize.”
Nature 630, 780-782 (2024)
doi: https://doi.org/10.1038/d41586-024-02029-w
This story originally appeared on: Nature - Author:Michael Eisenstein


















