Where did viruses come from? AlphaFold and other AIs are finding answers

Protein structures predicted by artificial intelligence have charted the evolution of the virus family responsible for dengue and hepatitis C
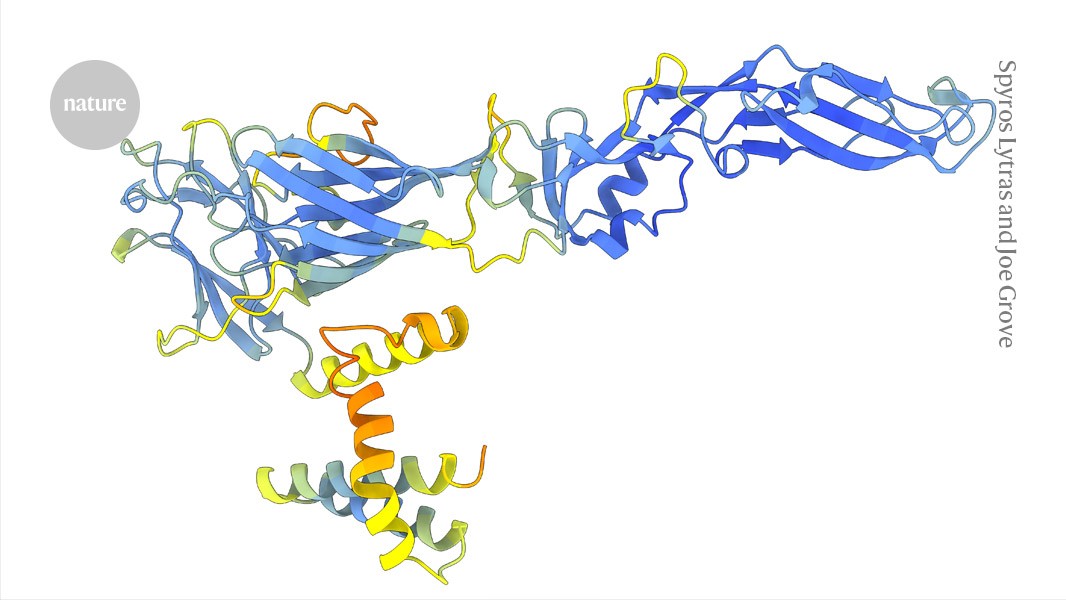
The structure of a dengue virus protein predicted using ColabFold–AlpahFold2.Credit: Spyros Lytras and Joe Grove
Artificial intelligence (AI) is helping to redraw the virus family tree. Predicted protein structures generated by AlphaFold and chatbot-inspired ‘protein language models’ have uncovered some surprising connections in a family of viruses that includes pathogens that infect humans as well as emerging threats.
Much of scientists’ understanding of viral evolution is based on genome comparisons. But the lightning-quick evolution of viruses — particularly those with genomes written in RNA — and their tendency to acquire genetic material from other organisms mean that genetic sequences can hide deep and distant relationships between viruses, which can also vary depending on the gene examined.
By contrast, the shapes, or structures, of the proteins encoded by viral genes tend to change slowly, which makes it possible to suss out these hidden evolutionary connections. But until the dawn of tools such as AlphaFold, which can predict protein structures at scale, it was not possible to compare protein structures across an entire viral family, says Joe Grove, a molecular virologist at the University of Glasgow, UK.
In a paper published this month in Nature1, Grove and his team demonstrate the power of a structure-based approach in the flaviviruses — a group that includes the hepatitis C, dengue and Zika viruses, as well as some major animal pathogens and species that could be emerging threats to human health.
How viruses enter
Much of researchers’ understanding of flavivirus evolution has been based on sequences of slow-evolving enzymes that copy their genetic material. However, researchers know remarkably little about the origins of the ‘viral entry’ proteins that flaviviruses use to invade cells and which determine the range of hosts they can infect. This gap, Grove argues, has slowed the development of an effective vaccine against hepatitis C, which kills hundreds of thousands of people each year.
“At the sequence level, things are so divergent that we can’t tell if they’re related or not,” he says. “The advent of protein structure prediction unlocks the whole question, and we can see things quite clearly.”
The researchers used DeepMind’s AlphaFold2 model and ESMFold, a structure-prediction tool developed at tech giant Meta, to generate more than 33,000 predicted structures for proteins from 458 flavivirus species. ESMFold is based on a language model trained on tens of millions of protein sequences. Unlike AlphaFold, it requires only a single input sequence, rather than relying on multiple sequences from similar proteins, so it might be especially useful for scrutinizing the most mysterious viruses.

A hepatitis C virus glycoprotein structure prediction. Credit: Spyros Lytras and Joe Grove
The predicted structures allowed the authors to identify viral entry proteins with very different sequences to those of known flaviviruses. They found some unexpected links. For instance, the subset of viruses that includes hepatitis C infects cells using a system similar to one they discovered in the pestiviruses — a group that includes classical swine fever virus, which causes haemorrhagic fever in pigs, and other animal pathogens.
The AI-enabled comparisons showed that this entry system is distinct from those of many other flaviviruses. “For hep C and its relatives, we don’t know where its entry system came from. It may have been ‘invented’ by those viruses way back when,” says Grove.
Stolen from bacteria
The predicted structures also revealed that the well-studied entry proteins of Zika and dengue virus have the same origins as those of what Grove describes as “weird and wonderful” flaviviruses with giant genomes, including Haseki tick virus, which can cause fever in humans. Another big surprise was the discovery that some flaviviruses have an enzyme that seems to have been stolen from bacteria.
“This would be unprecedented,” says virologist Mary Petrone, at the University of Sydney, Australia, were it not for her team’s discovery this year of a similar theft in an especially weird and wonderful flavivirus species2. “Genetic piracy could have played a larger role in shaping the evolution of the flavivirids than previously thought,” she adds.
David Moi, a computational biologist at the University of Lausanne, Switzerland, says that the flavivirus study is the tip of the iceberg, and that the evolutionary histories of other viruses and even some cellular organisms are likely to be rewritten with AI. “We’ll be retelling their stories with a new generation of tools,” he says. “Now that we can see a bit farther, all of these things are going to have to have a little bit of an update.”
doi: https://doi.org/10.1038/d41586-024-02970-w
This story originally appeared on: Nature - Author:Ewen Callaway


















