Can AI build a virtual cell? Scientists race to model life’s smallest unit

Several groups hope to develop artificial-intelligence models that can predict how cells behave
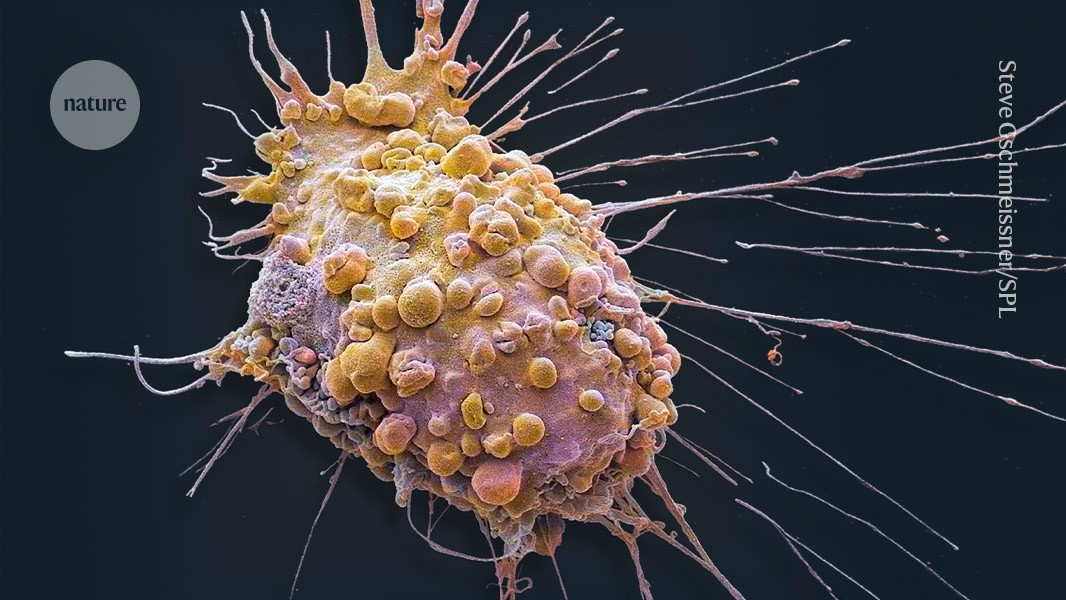
Virtual cell models could help to predict how tumour cells (pictured) respond to experimental drugs. Credit: Steve Gschmeissner/Science Photo Library
If Stephen Quake gets his way, biologists in the future will spend a lot less time wielding pipettes. “Our goal,” he says, “is to create computational tools so that cell biology goes from being 90% experimental and 10% computational to the other way around.”
Quake, head of science at the Chan Zuckerberg Initiative (CZI) in Redwood City, California, is among the researchers leading a charge to create virtual cells. These are artificial intelligence (AI) models that can generate the kinds of insight that currently take weeks of experiments to answer — such as how tumour cells will respond to a particular drug.
“It’s going to be a very powerful tool for understanding what goes wrong in disease,” says Quake, who envisions that scientists will use experiments mainly to validate predictions made by virtual cells.
Efforts to create virtual cells are in their early days, but the idea has attracted intense interest in both academic and industry laboratories worldwide. CZI, a non-profit organization that develops open data sets and tools, is planning to spend hundreds of millions of dollars on creating virtual cells over the next decade. Google DeepMind, in London, has a virtual-cell project too, its chief executive Demis Hassabis said earlier this year.
“This is a gigantic task,” says Jan Ellenberg, a molecular biologist at the Science for Life Laboratory, a national research organization in Solna, Sweden. He is co-leading the lab’s own virtual-cell model called Alpha Cell, which will launch in 2026. “What is possible now and needed now is to have the first pioneering projects that show this can, in principle, work.”
But some scientists say that the rush to develop virtual cells — although an important long-term goal for biology — hast a lot of hype, but not a lot of concrete results or a clear path to success.
“It’s primarily being used as a rallying cry and a funding mechanism, and it’s working,” says Anshul Kundaje, a computational biologist at Stanford University in California. “Investors are putting huge amount of funding into this space.”
Bugs in the machine
Biologists have been using computers to model cellular behaviour for decades. In 2012, scientists created the first computational model of an entire cell, capturing the inner workings of the bacteria Mycoplasma genitalium, which has just 525 genes1.
But these and other early efforts “were often trying to really build a full mechanistic model of the cell”, says Silvana Konermann, a computational biologist at the Arc Institute in Palo Alto, California.
By contrast, the current push to develop virtual cells takes advantage of advances in AI that can develop sophisticated representations of data, such as text in the case of large language models, when fed vast quantities of it. “Building models that learn from data is revolutionary,” says Quake.
Early virtual-cell offerings have focused largely on one type of data: those from experiments that sequence all the messenger-RNA molecules in individual cells, amounting to a catalogue of gene activity and a snapshot of the cell’s current state.
These data form the basis of ‘atlases’ that map different cell types across humans and other organisms, revealing underappreciated diversity. Researchers are now churning out ‘single-cell sequencing’ data sets to help power their virtual cells. CZI has plans to release sequencing data from one billion cells (expanding a database of more than 100 million), and in February, Arc released sequencing data from 100 million cancer cells treated with hundreds of drugs.
Single-cell sequencing data is appealing, says systems biologist Hani Goodarzi at the Arc Institute, because it can be affordably generated at a similar scale to that at which large language models start to gain sophisticated capabilities — in the hundreds of billions of datapoints.
Login or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Sign in or create an accountdoi: https://doi.org/10.1038/d41586-025-02011-0
This story originally appeared on: Nature - Author:Ewen Callaway


















